Making the Prenomics summary poster
I recently designed a poster that acts as a “cheat sheet” for Cloud-SPAN’s Prenomics course. Here I’ll give a quick overview of my thought process while making the poster and how I chose what to include.
Command cheatsheet
I started with the command cheatsheet, which is a bit back-to-front given that the command line comes chronologically last in the course. However, it seemed like an easy entry point. I had previously produced a glossary for Prenomics, so I took this and organised the commands into logical groupings based on their function.
I ended up with four groups: navigating files, viewing files, editing files and searching files. There were two commands which didn’t fit into these groups, which were history and man. I tried to find a way to include them, but ultimately left them out as I decided they were not crucial knowledge.
I tried to format the commands in a way that made it clear how to use them while still keeping it general and easily readable. I used colour to clarify which parts of the command corresponded to which parts of the explanation. For example:

In this case ‘file’ (the file to be moved) is pale blue while ‘directory’ (the location to which the file should be moved) is dark blue. I also used underlining to indicate where the command came from - in this case, mv comes from the m and the v of ‘move’. My aim here was to aid recall of the command in future, as hopefully it will help the reader make a stronger mental connection between the command mv and its function (‘move a file’).
Files, paths and file types
The command line makes up a significant portion of the Prenomics course, but it could be summarised in a relatively small amount of space. To work out what else should be included, I looked at the course more holistically. The first lesson of the course is about files and directories, so it was clear to me that there needed to be a section about this. This lesson also covers the file types used in the course, including .FASTQ and .PEM files which are likely to be new to most learners, so I wanted to include this too.
I found it difficult to summarise the information about file paths and directories into a mostly graphical format. In particular, I found that I couldn’t rely on giving examples as much as we do in the course itself, due to the need to reduce text as much as possible. I ended up just including definitions of absolute and relative paths, along with a diagram of the file system inside the Cloud-SPAN AWS instance.
If I’d had space, I would have included more information on working directories and examples of how the file system diagram can be represented with file paths.
Why use…?
The next sections I designed were the ‘Why learn command line?’ and ‘Why use the cloud?’ sections.
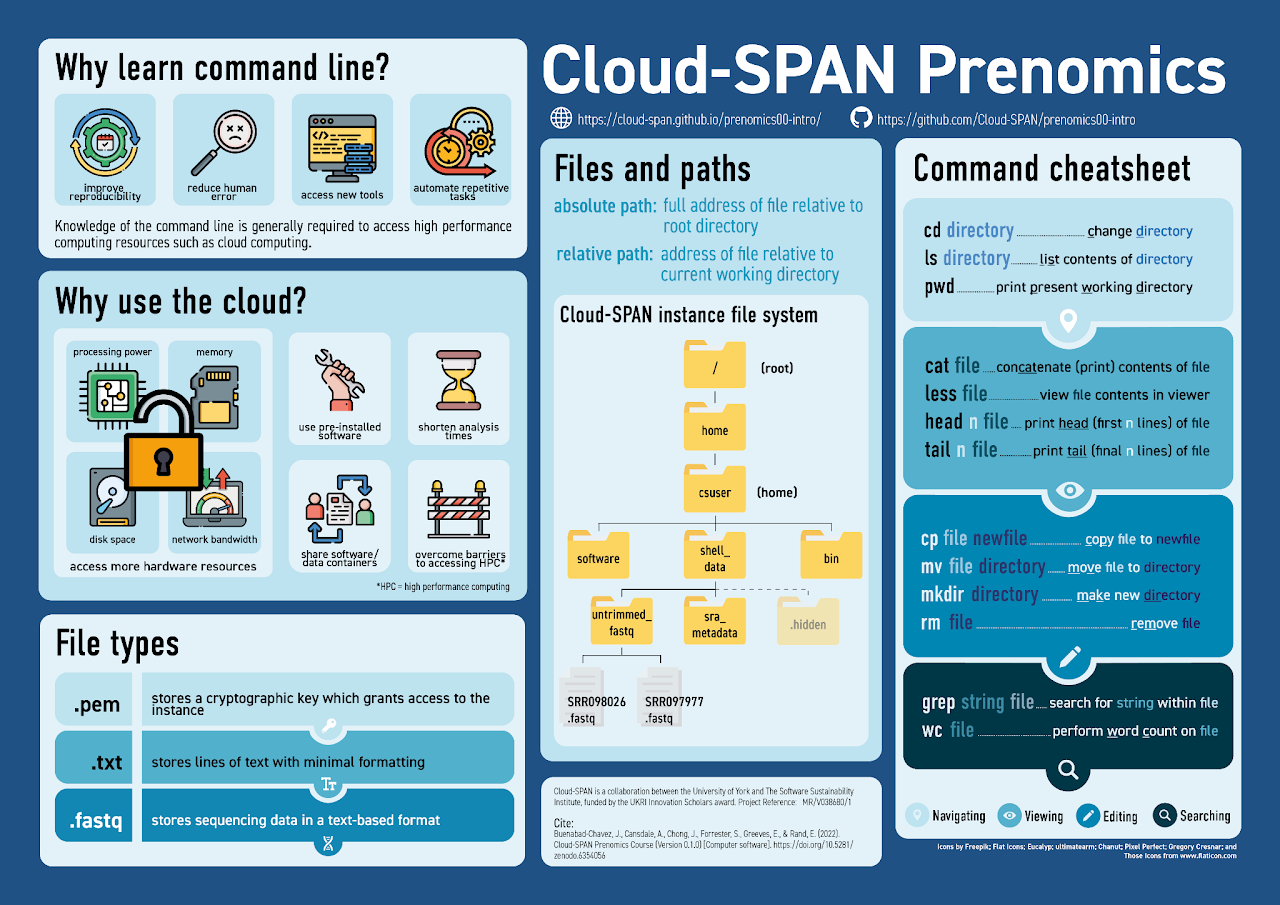
The question of why the command line is useful is covered in episode three of the first day of Prenomics, as part of the introduction to the shell. I distilled the reasons given here down into four main themes: automation of repetitive tasks, reducing human error (as a result of automation), improving reproducibility and the ability to access new tools (either because the command line offers more functionality or because it opens up use of high performance computing systems (HPC) such as the cloud).
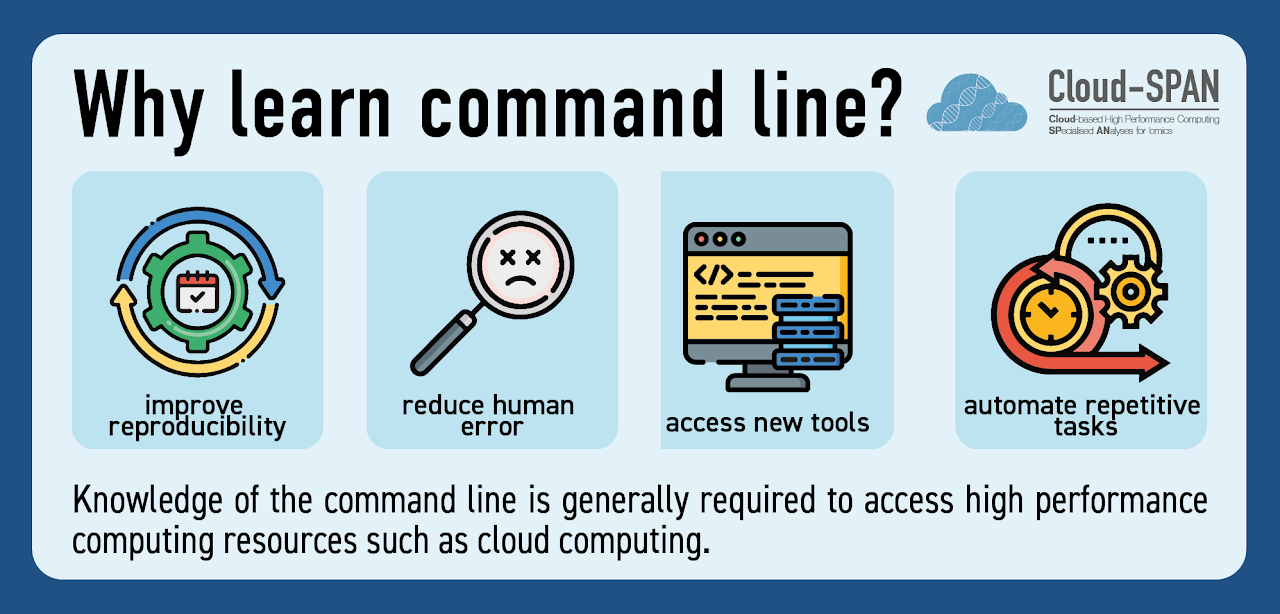
The other question- why cloud computing is useful- is not technically covered in Prenomics material. However, it is discussed in our Genomics course, where the three reasons given for using HPC are lack of resources needed to run analyses, analyses taking a long time to run and problems installing software. These reasons align closely with three of the challenges identified by Cloud-SPAN project lead, James Chong, as facing the field of metagenomics (hardware, time and software).
However, these reasons could actually be used as a reason to use any kind of HPC resource, not just the cloud. I wanted to include reasons specific to the cloud. The two major reasons I came across were the ability to share software or data containers across different institutions, and use of the cloud when other HPC is inaccessible.
In an earlier version of the poster, the ‘why use cloud?’ section was framed in terms of challenges that users might face, such as long analysis times or issues installing software. I reworded this section to match the framing of the ‘why learn command line?’ section; that is, a solutions-oriented summary, with ‘shorten analysis times’ replacing ‘long analysis times’ and ‘use pre-installed software’’ replacing ‘issues installing software’.
File types
Lastly I wanted to include a brief summary of the file types used in the Prenomics course, as it is likely that two out of three of these will be new to learners. This part was quite easy - I just wrote a short sentence to describe each file type and paired it with the relevant file extension.
In the course a significant amount of time is dedicated to introducing the .fastq file structure, which codes sequencing data into a text format with four lines per read. I considered including this information but I didn’t have room.
The big reveal…
And finally, here’s the full poster! You can download a high-res version of the poster for your own enjoyment here.